Getting Started
 Derailment (Ray's Academic Blog)
Derailment (Ray's Academic Blog)
Derailment has posts on how to set up a Mac:
There are also posts on how to set up a Windows machine:
There are posts on configuring and updating:
Why Bother?
The Gold Standard
- Data scientists can archive a compendium of work, which includes the raw data, all the data processing/analyses code, all the analysis software itself and the resulting manuscript in a bundle.
This perfectly reproducible workflow requires extra R packages likerenvand extra software likedocker.

- This also requires extraordinary care to deal with Protected Health Information.
Good Work
- Non-experts can easily build a project which will knit together all of the data processing and the text, tables and figures for a paper into a single document and share that information with the world.
This workflow, which excludes the raw data and the analysis software itself, goes a long way toward allowing reproduction of findings.
Where We are Going
- Use modern R and RStudio to quickly and easily set up a paper outline/shell for a biomedical research article.
- Easily organize a research project and embed analysis, figures, and tables directly into your article.
- Stop copying and pasting analysis/tables out of Excel into Word.
- You get the shell for a paper in just a couple clicks.

Just how quick and easy? Just add rUM...
- Click to install a package.
- Type rUM. (capitalization matters)
- Restart RStudio.
- Click New Project... from the File menu.
- Click New Directory.
- Click Research Project Template.
- Choose where to save the project.
- Click Knit.

Demo
What does that hard work  get you?
get you?
The rUM project template creates a folder that contains:
- The paper shell, called reproducible.Rmd (you can rename it)
- An empty folder, called data, to hold your data
- An RStudio project file, with the name you chose
- Bibliography files, named references.bib, to hold references to articles and an automatic bibliography file, named packages.bib with references to all the used R packages
- A bibliography style file, named the-new-england-journal-of-medicine.csl to format your references
- A file, named .gitignore to help protect you from accidentally sharing data
Authoring
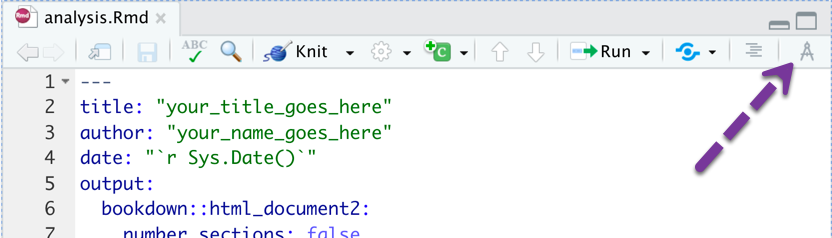
How Do I Write/Edit in RStudio?
The document is in a format called R Markdown. It saves with a suffix of .Rmd. You can use point-and-click tools to edit it.
Click the Visual Markdown Editor button:
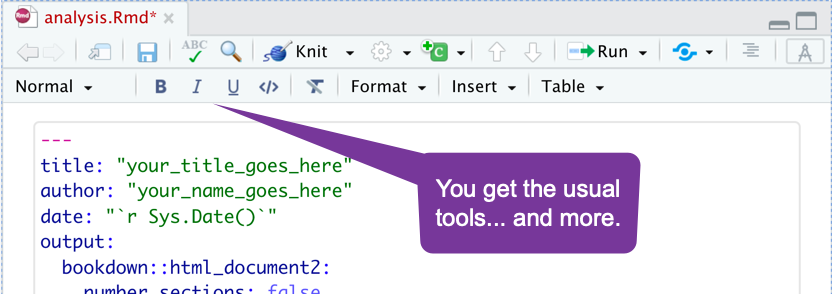
Editing Tools - Formatting Text
It works like a word processor. Highlight and click a button.
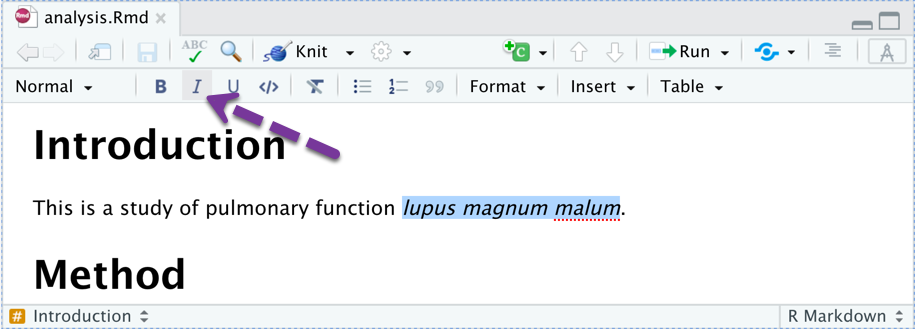
Editing Tools - Spelling
Right click to fix spelling or add to the dictionary:

Navigation
You can jump to sections using the document outline
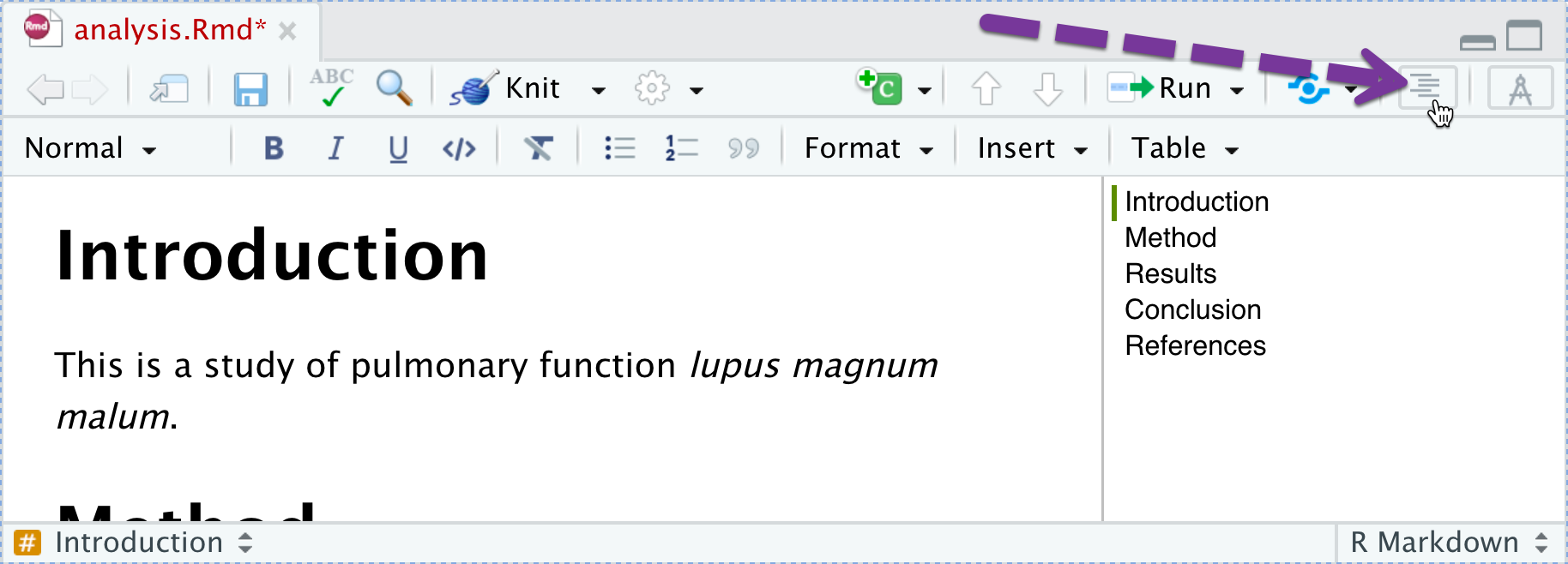
Navigation
You can jump to sections using the document outline

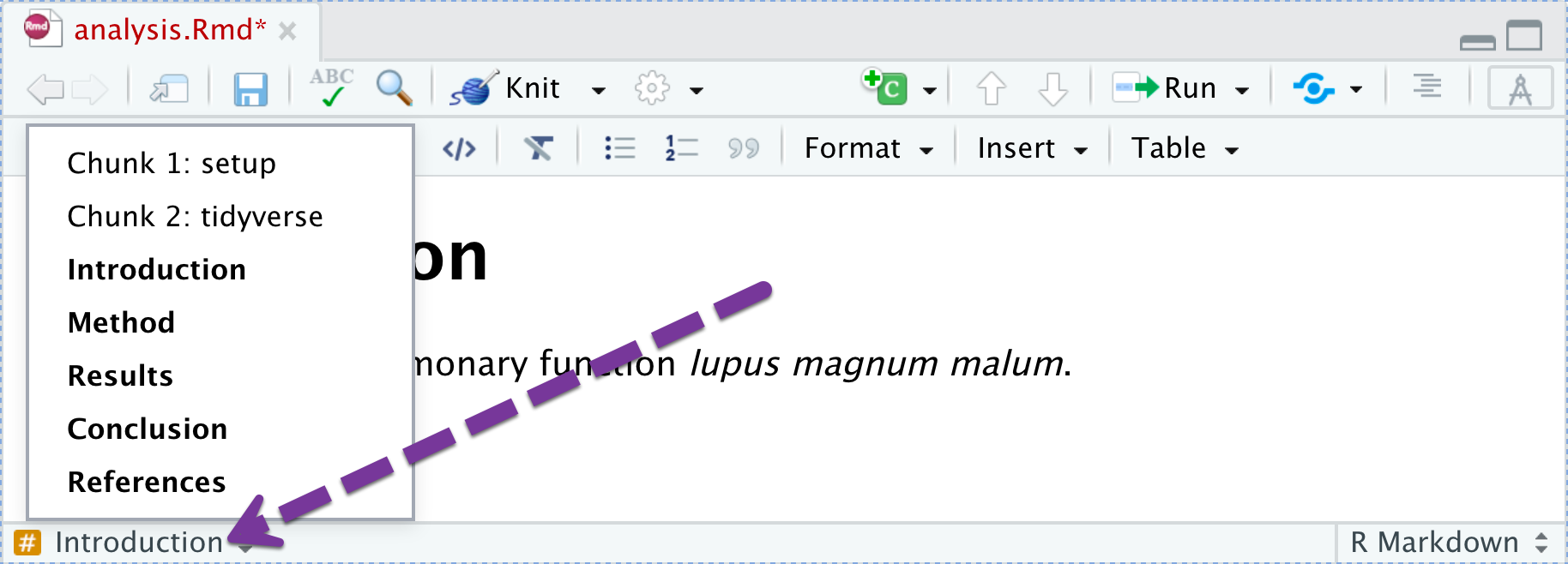
Cross Referencing (1)
You can add IDs to sections of your paper (as well as figures/tables) and then put in hyperlinks. To add a section identifier:
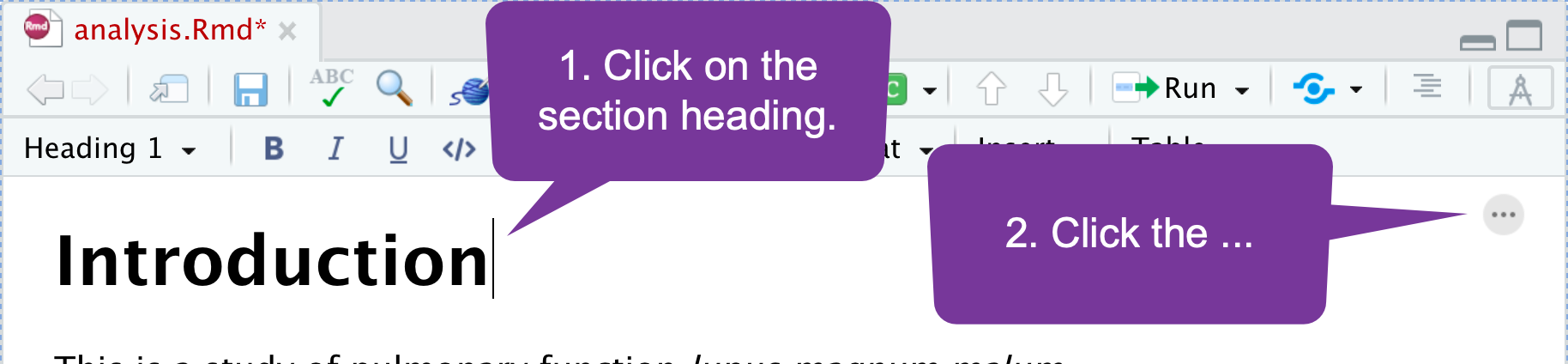
Cross Referencing (1)
You can add IDs to sections of your paper (as well as figures/tables) and then put in hyperlinks. To add a section identifier:

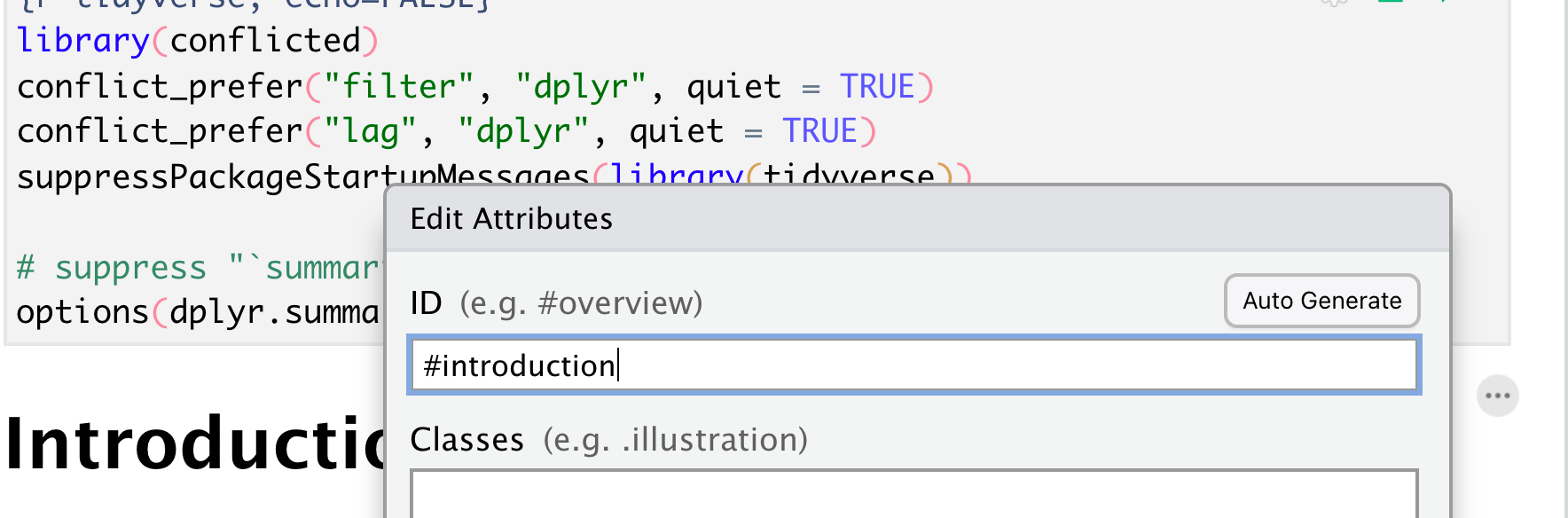
Cross Referencing (1)
You can add IDs to sections of your paper (as well as figures/tables) and then put in hyperlinks. To add a section identifier:



Cross Referencing (2)
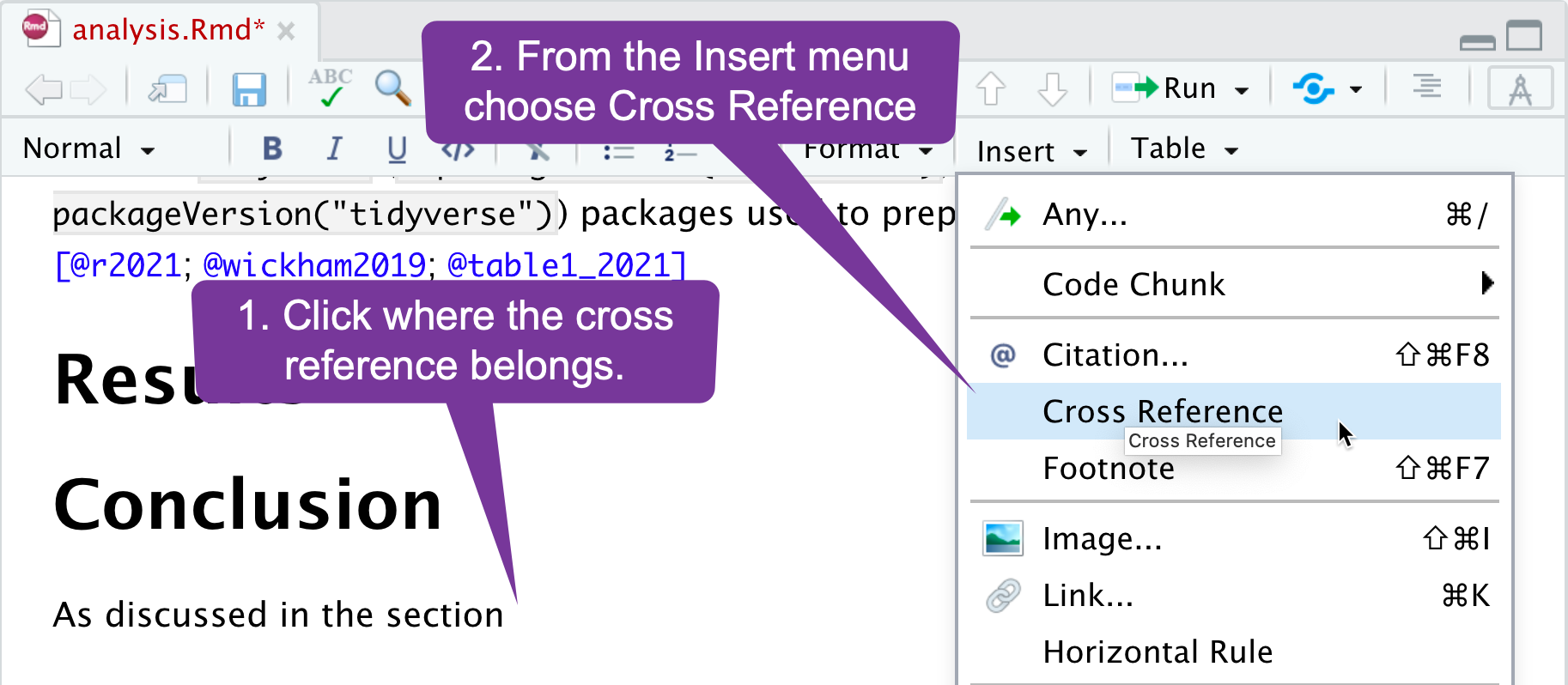
If your paper is using section numbering, you can use the menu to add links to sections:
Cross Referencing (2)
If your paper is using section numbering, you can use the menu to add links to sections:

Cross Referencing (2)
If your paper is using section numbering, you can use the menu to add links to sections:



Cross Referencing (3)
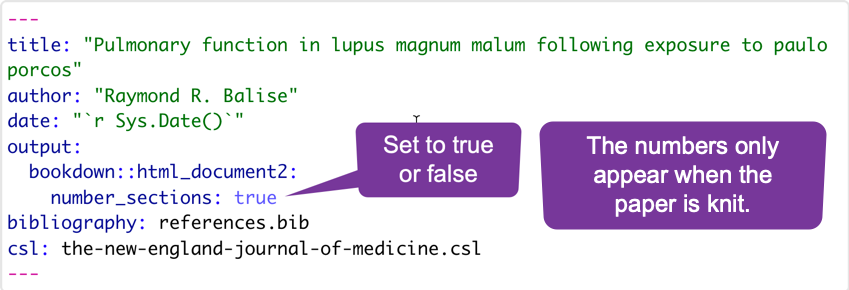
If your paper is not using section numbering, you need to type some markdown code. The syntax to cross reference a section is:
[label](#link)where the label is the text for the reader and #link is the section ID.
Cross Referencing (3)
If your paper is not using section numbering, you need to type some markdown code. The syntax to cross reference a section is:
[label](#link)where the label is the text for the reader and #link is the section ID.

Cross Referencing (3)
If your paper is not using section numbering, you need to type some markdown code. The syntax to cross reference a section is:
[label](#link)where the label is the text for the reader and #link is the section ID.


Cross Referencing (3)
If your paper is not using section numbering, you need to type some markdown code. The syntax to cross reference a section is:
[label](#link)where the label is the text for the reader and #link is the section ID.



Doing Calculations in Text
You can nest a calculation directly into the narrative by clicking the code button.
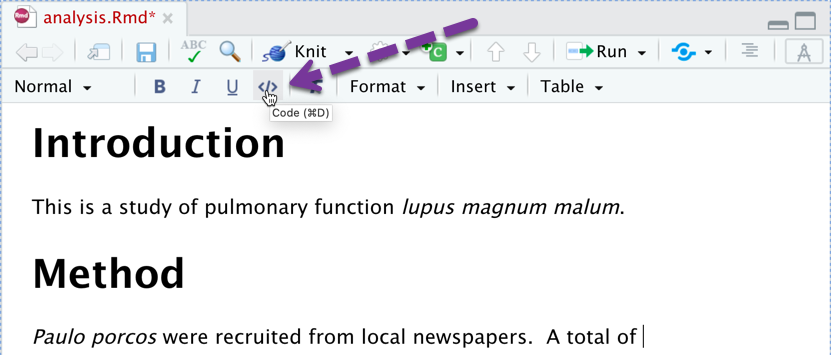
Doing Calculations in Text
You can nest a calculation directly into the narrative by clicking the code button.


Preprocessing Code Blocks
If you need to do something complicated, like calculating how many subjects were enrolled, you can add a code block. You can do some calculating, then reference the results.
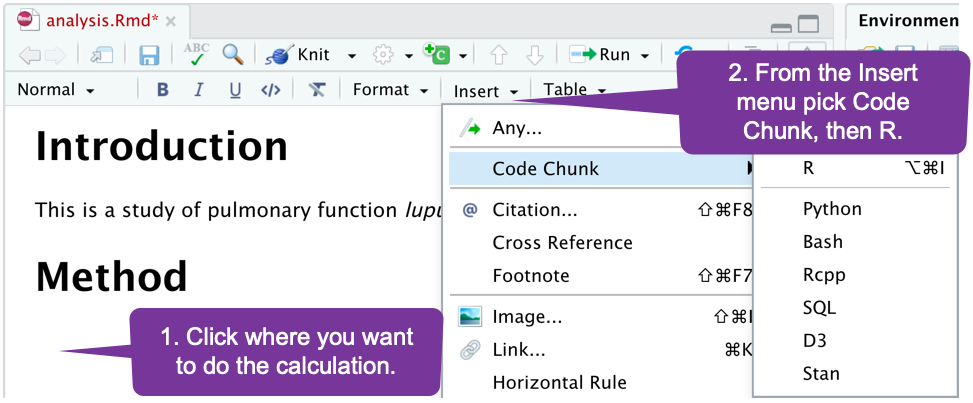
Using a Code Block
Write your R code below the line with {r}. You can later reference any object you have created.
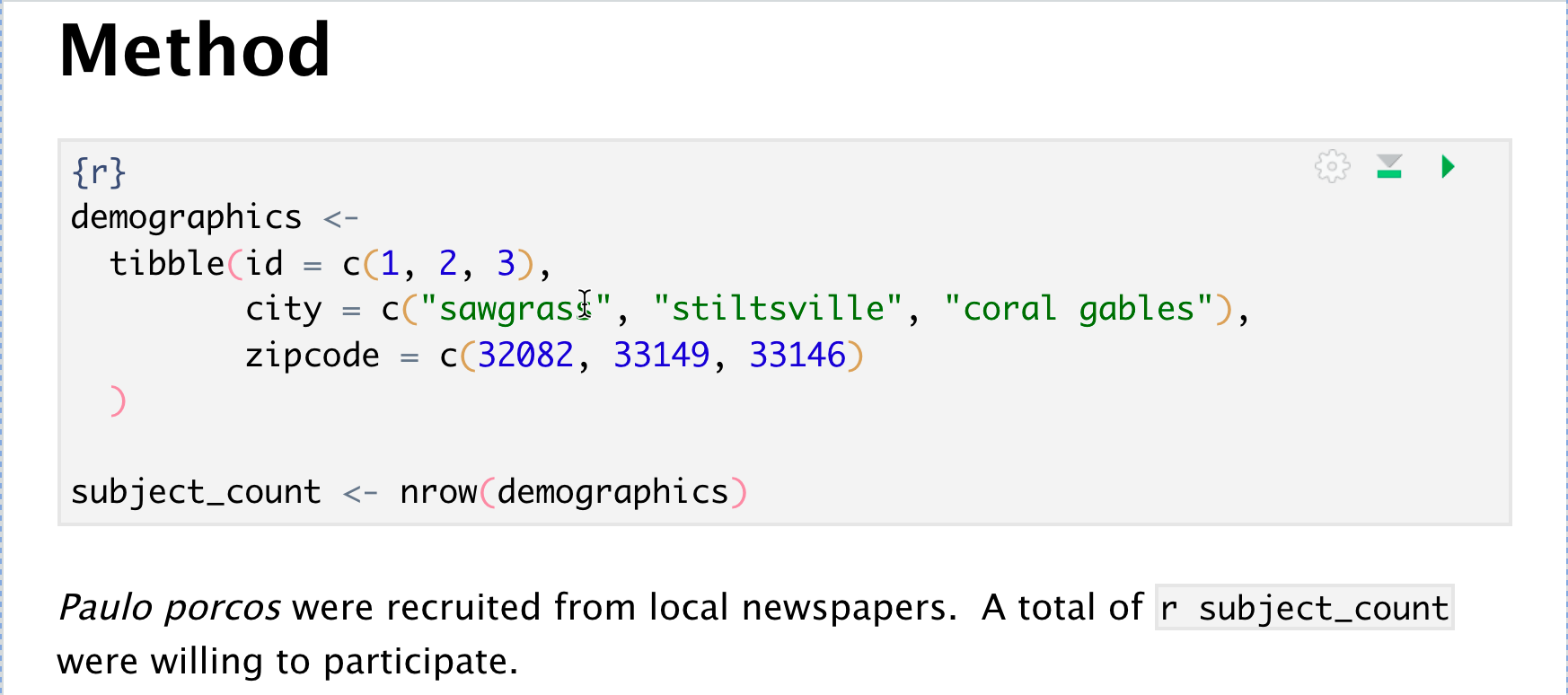
Using a Code Block
Write your R code below the line with {r}. You can later reference any object you have created.

And you can use functions in inline code.
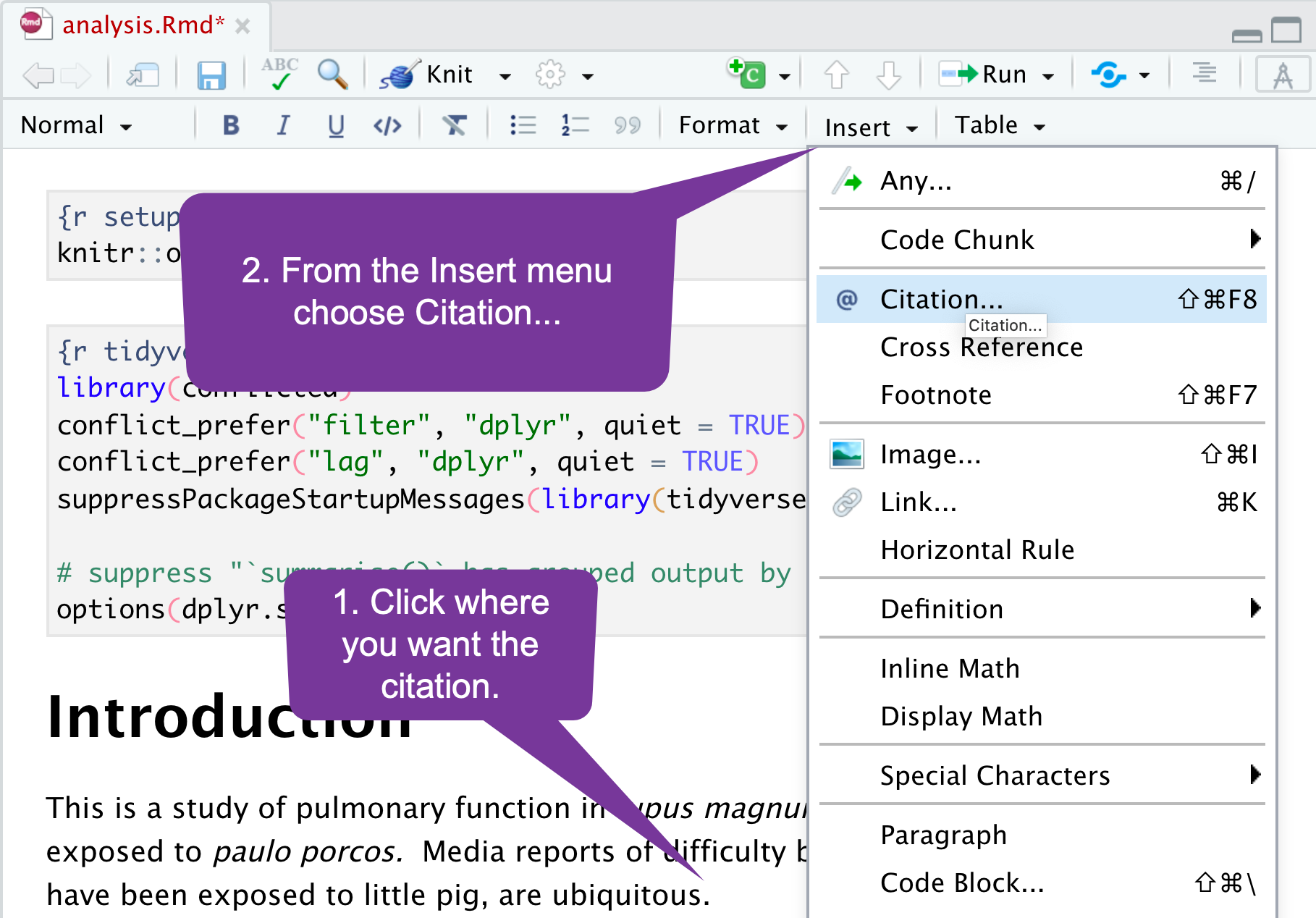
Adding References (1)
Adding references/citations is simple:
Adding References (2)
There are many search engines to help you find your article:
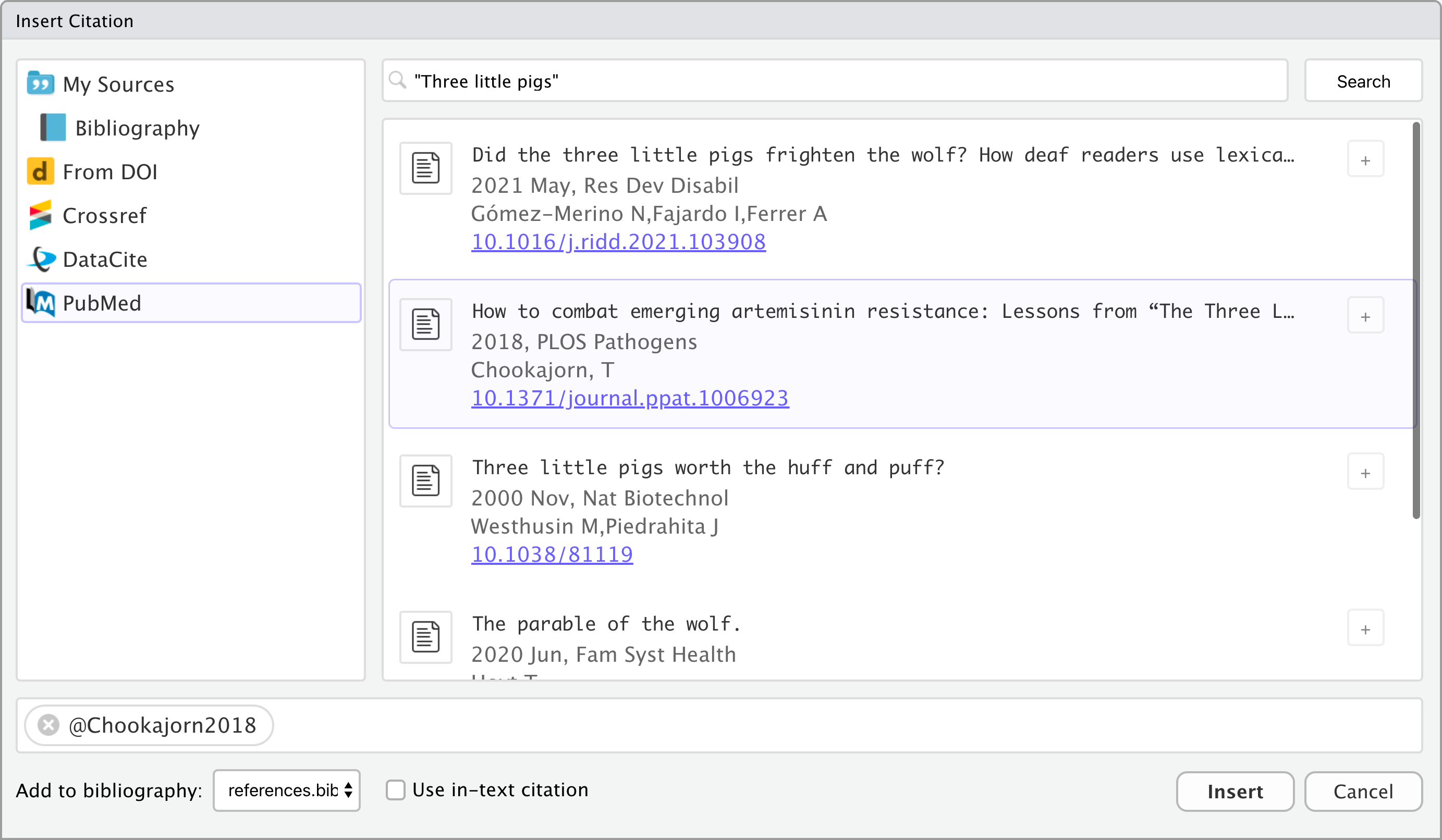
Adding References - NEJM Style
New England Journal citations go at the end of a sentence.

The output appears as:

To Use a Different Bibliography Style
Search the Zotero Style Repository website and save the ".csl" file into your project folder.
Then change the YAML header in the ".Rmd" file to match the file you downloaded:
---title: "your_title_goes_here"author: "your_name_goes_here"date: "r Sys.Date()"output: bookdown::html_document2: number_sections: falsebibliography: [references.bib, packages.bib]csl: the-new-england-journal-of-medicine.csl---Using American Psychological Association (APA) Style
If you download the apa.cls file into the project folder and tweak the YAML header, your references will appear in APA format.
End of Sentence References - use [ ]

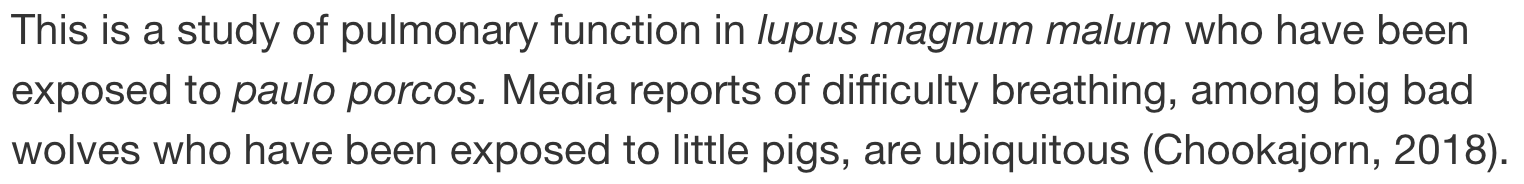
Inline References - without [ ]


Using GitHub
Why Bother?
- To get a job...
- Anybody who claims to do data science or who wants a job in industry needs to know how to use git/GitHub.
- If somebody does not have GitHub on the top of their resume I stop reading.
- I look at GitHub usage to judge if I want to hire somebody.
- Most people in the R world post code on GitHub. You can see how other people solve problems.
- If you do something useful you can share.
- It is nearly no effort to share your algorithms with the world.
- You can also do pretty hardcore project management with GitHub.
Using git and GitHub
GitHubis a website that lets you share any file with the world.gitis software that helps you keep files synchronized and helps resolve conflicts if more than one person has edited the same part of a file.- Derailment has a post to help you set up
gitandGitHub. - Obviously you need to be careful that you do not put sensitive data like Protected Health Information (PHI) and Personally Identifiable Information (PII) onto GitHub.
- If you build a project using
rUM, it automatically puts a file into the project folder, called.gitignore, which helps block the sharing of files that it recognizes as data.
How to Make a Project on GitHub
- Log into your account.
- Create a new project.
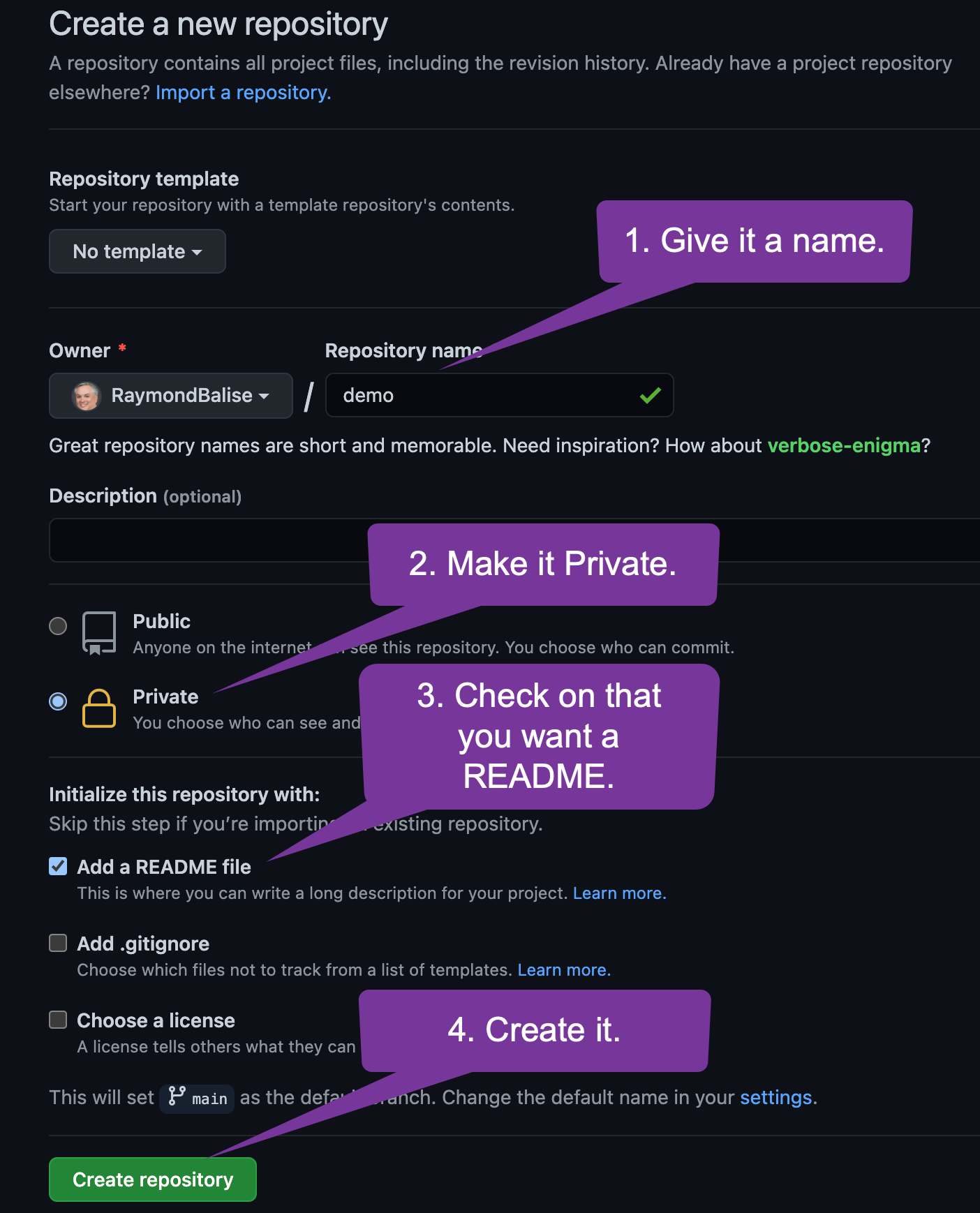
Get the URL so you can sync your local machine.
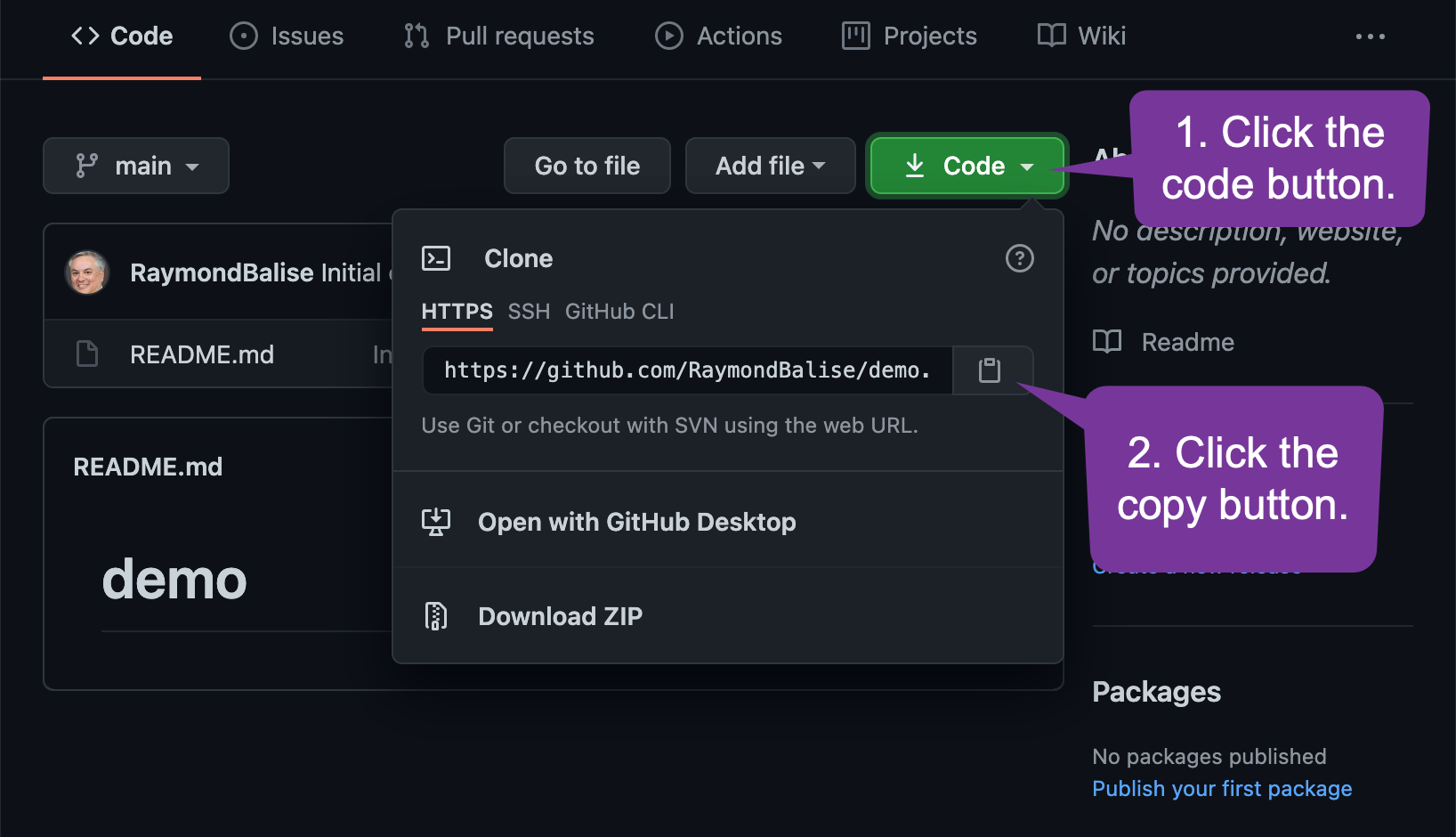
To Create a Local Copy of the Files that are on GitHub
- Open RStudio
- Use the File > New Project ...
- Choose "Version Control"
- Choose "Git"
- Add your project URL:
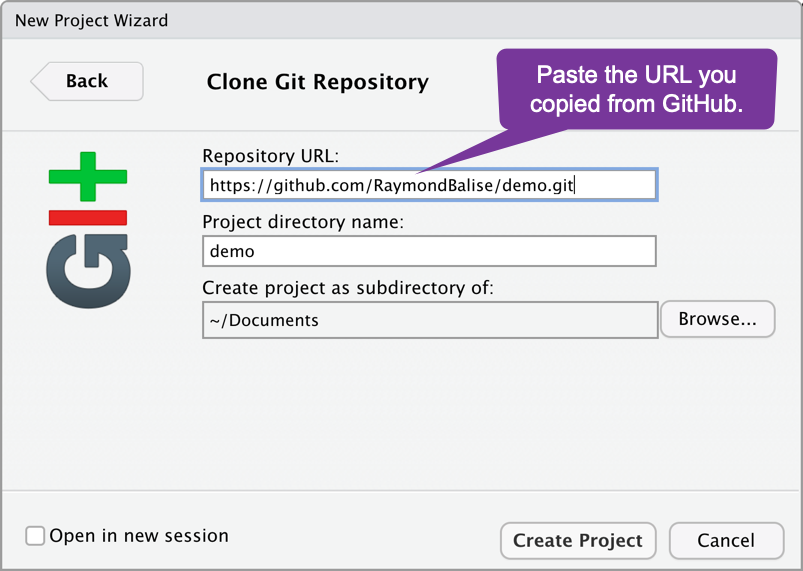
The Git Enabled Project
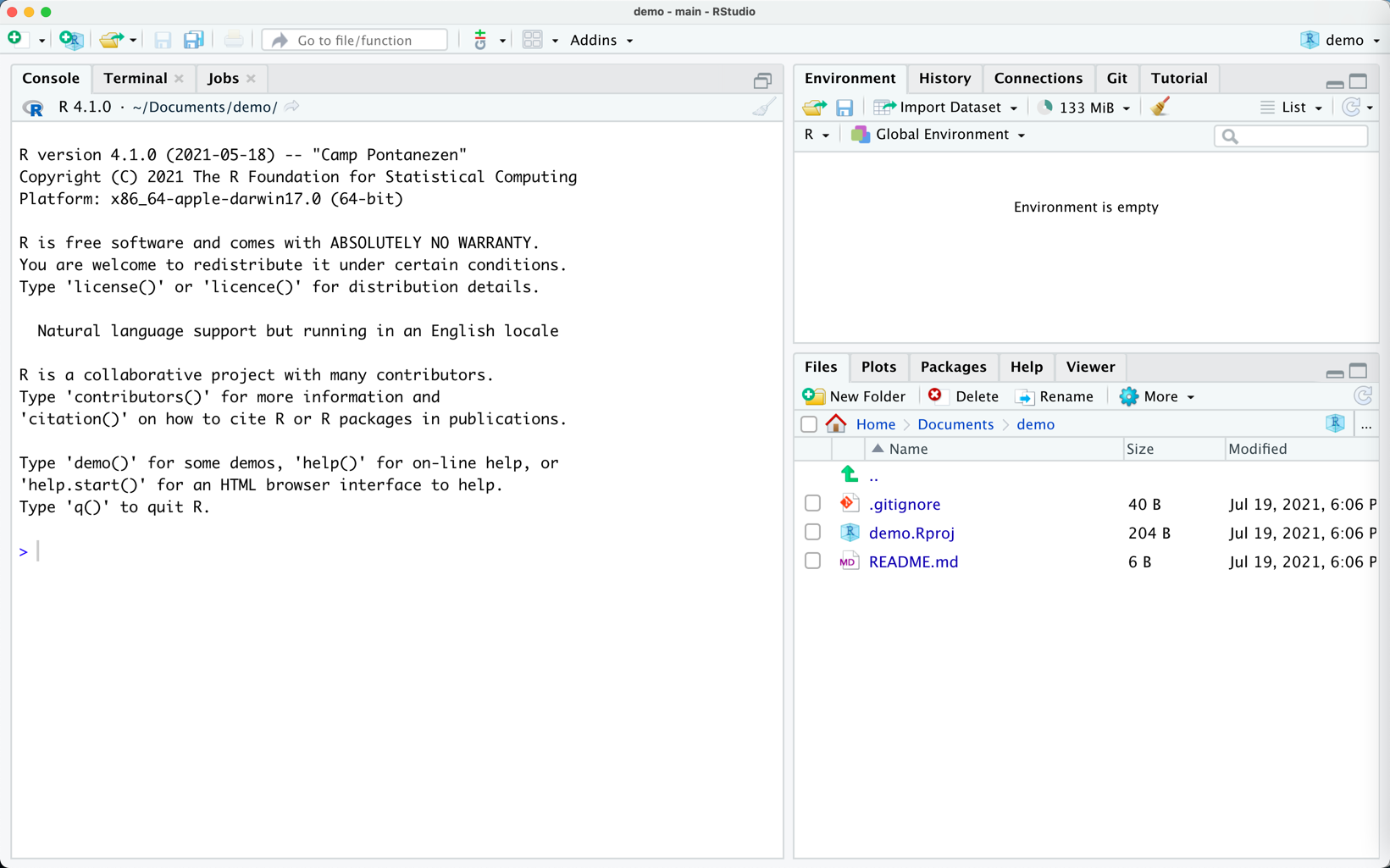
Notice the Files and the Git Tab
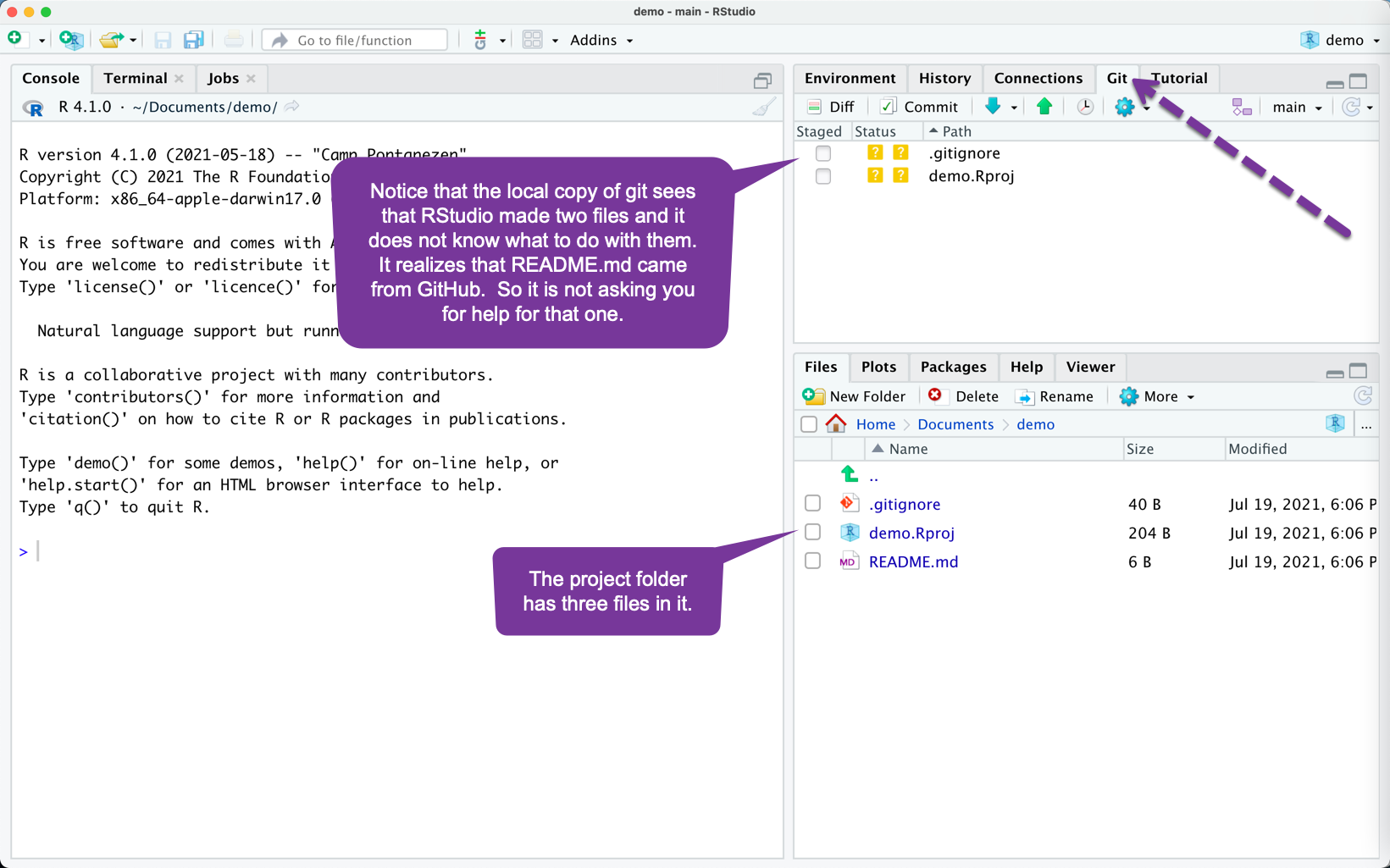
If You Have a Project Created with rUM (1)
You can copy the contents of the rUM folder into this GitHub enabled project folder but this requires a little care....
- Using RStudio, delete the file called
.gitignorethat was automatically created in the GitHub enabled project.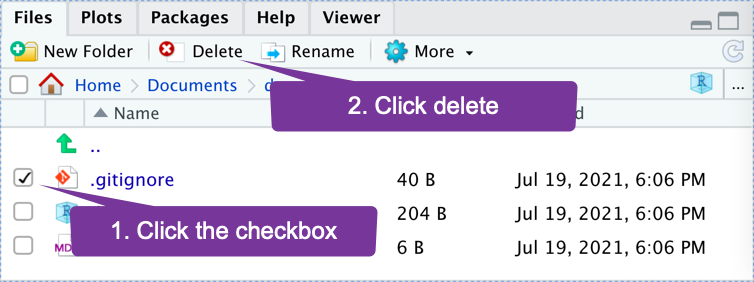
If You Have a Project Created with rUM (2)
Move the content of your rUM project folder into the GitHub enabled project folder.
- Windows
- Using the Windows File Explore, select all files in the rUM project, then copy and paste the files into the GitHub enabled project. Tell it to overwrite the README file.
- Mac
- You need to move the
.gitignorefile (which is hidden) along with everything else. - Show hidden files before you move files. To show hidden files in the Mac Finder, hold down the command key (it looks like ⌘) and the
shiftkey and then tap the.(period/decimal) key. - After you move the .gitignore file, you can do
⌘ shift .again to hide the hidden files.
- You need to move the
Dealing with Hidden Files on Mac
- Files whose names starts with a
.(period/decimal) are hidden on Mac.By default, the RStudio IDE shows some hidden files:
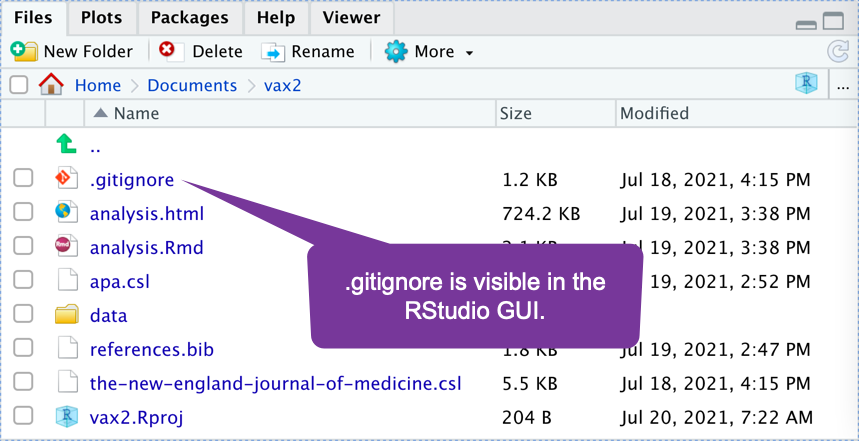
By default, the finder hides all hidden files:
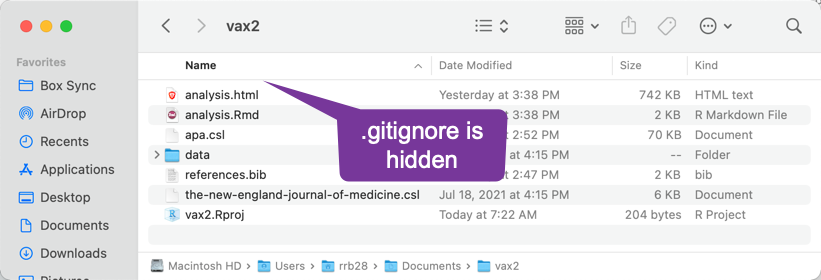
Show Hidden Files on Mac
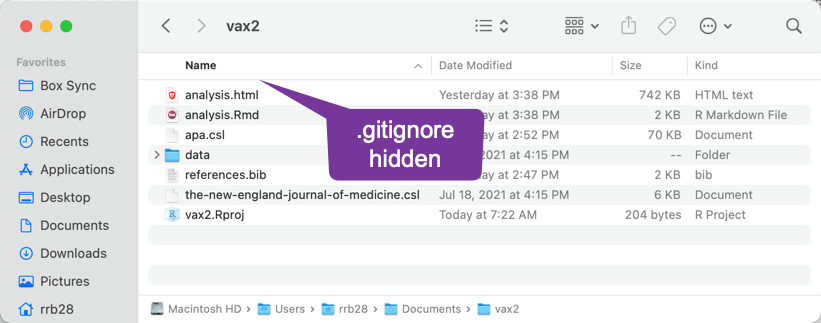
- Show hidden files with
⌘ shift .The finder hiding hidden files:
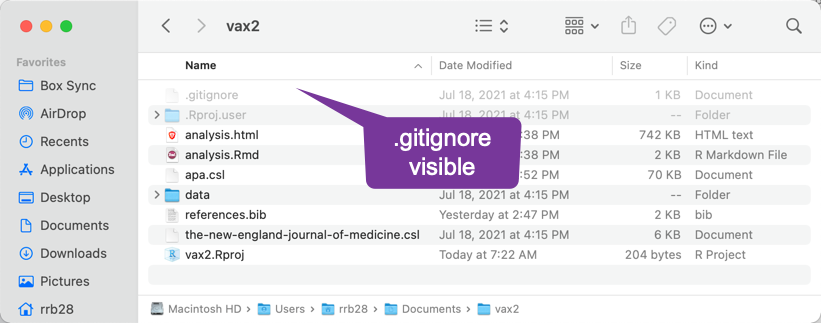
The finder showing hidden files:
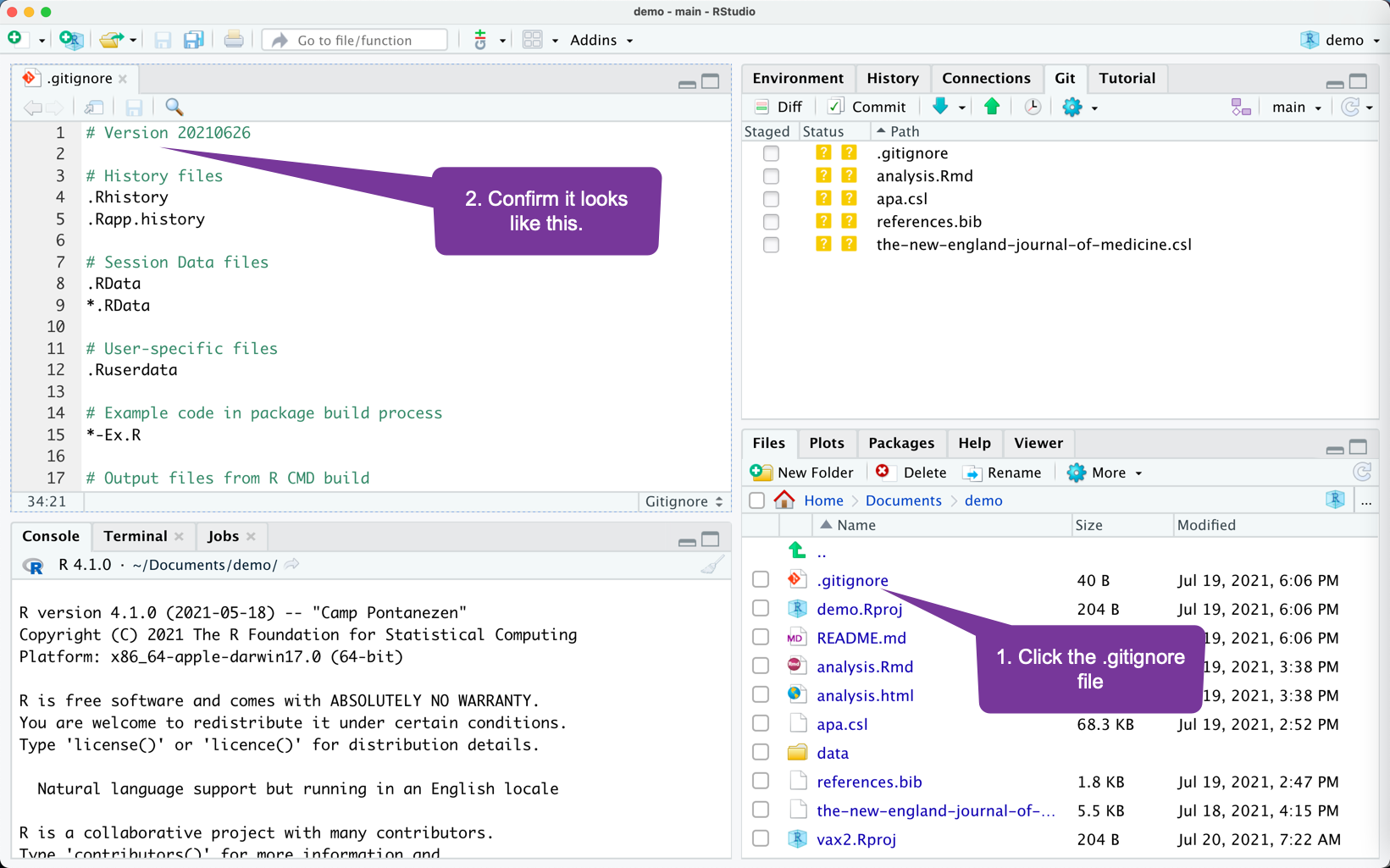
After Moving Files to the GitHub Enabled Project
What is the big deal about .gitignore?
- Notice the set of files that are listed in the Git windowpane.
- These are files that can be tracked/backed-up/sent to GitHub.
- The .gitignore specifies files/folders to not track/back-up/sent to GitHub.
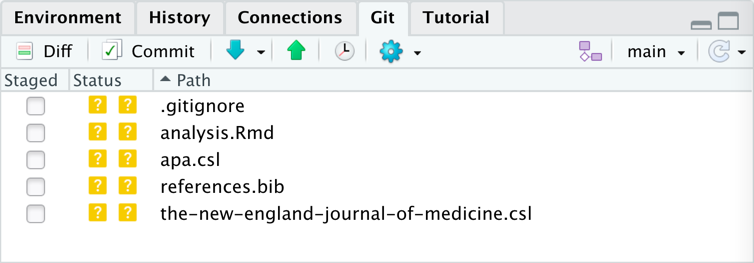
- My
.gitignorefile lists data files that I work with. If you work with other kinds of files that contain data, tell me and I will add them to my list.
.Ray's .gitignore
- My
.gitignorefile lists data files that I work with. - If you want to get the latest version without using
rUM, go here: https://gist.github.com/RaymondBalise

How do I move a file to GitHub? (1)
- You choose which files.
- Add a message to describe the work.
- Send it.
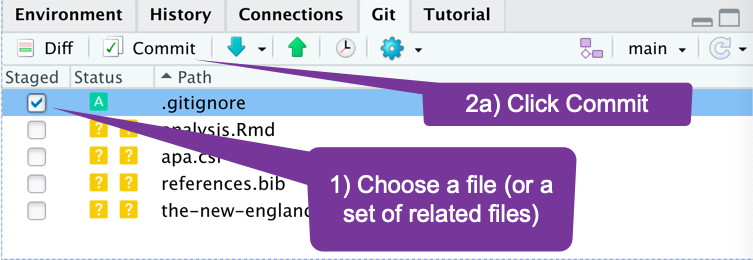
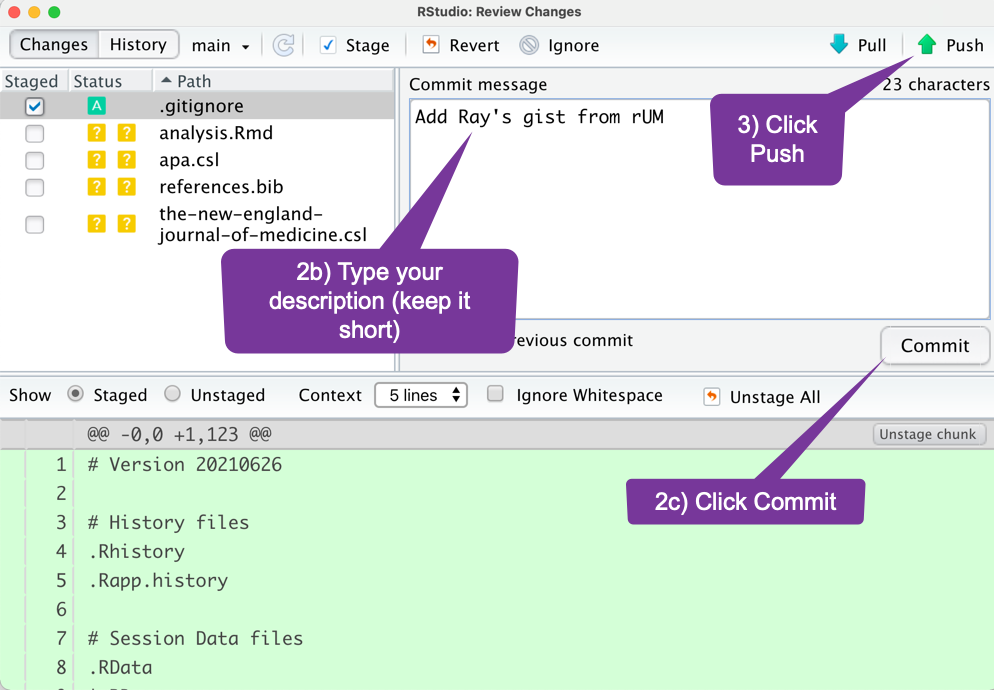
How do I move several related files to GitHub? (2)
- You choose which files.
- Add a message to describe the work.
- Send it.
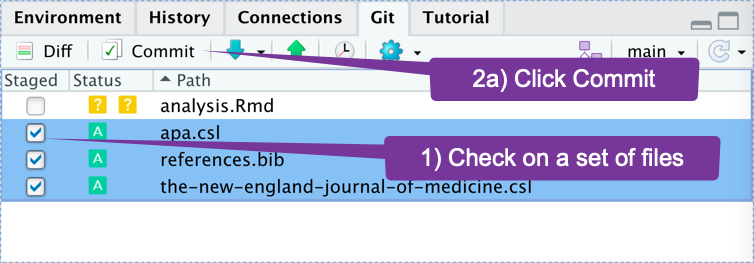
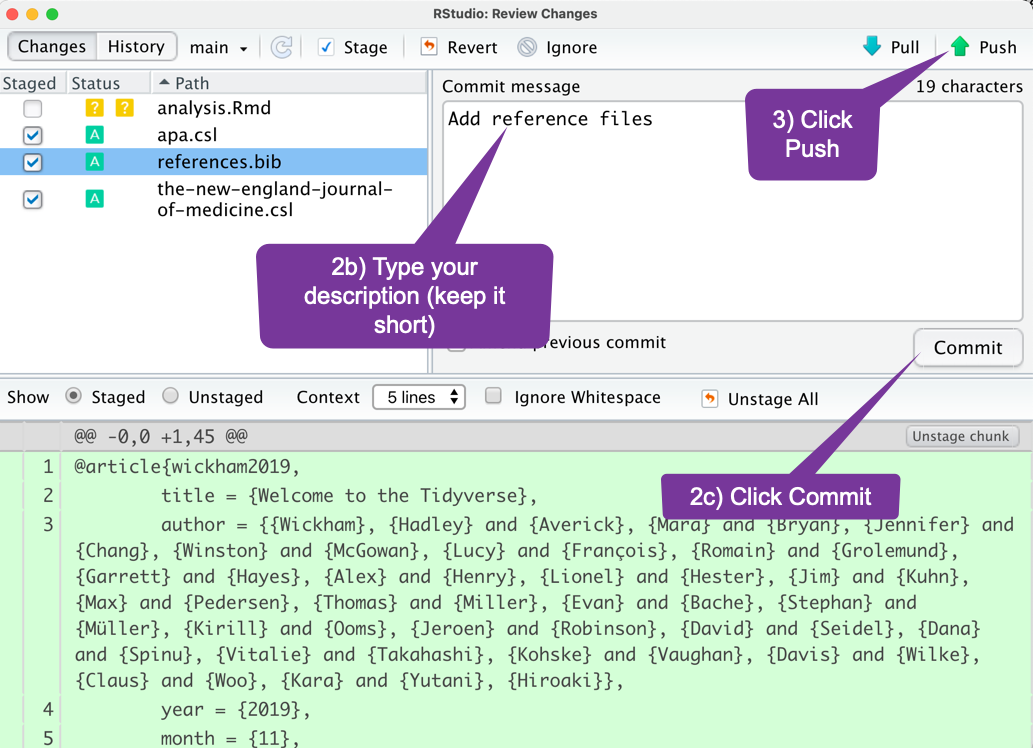
If You Want to Intimidate People
- You choose which files. Call this "staging files."
- Add a message to describe the work. Call this "committing files."
- Send it. Call this "pushing files."
Call your git/GitHub project back-up/repository/web-page a repo.
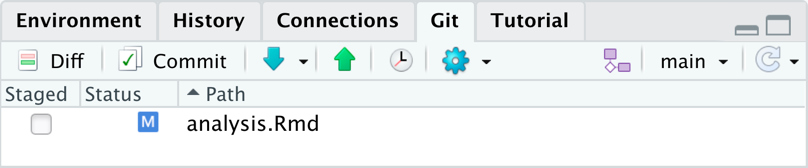
Modified Analysis File
- When you save and commit a file, it disappears from the
gitwindowpane. - As soon as you modify a file, that you previously committed it it will be listed again in the
gitwindowpane.- It will appear with a blue M to indicate the file was modified.
- When you are happy with your modifications, upload it to GitHub using the same workflow.
To use the git terms:
- Save
- Stage (check off the box)
- Commit with a message (type a summary)
- Push (upload)
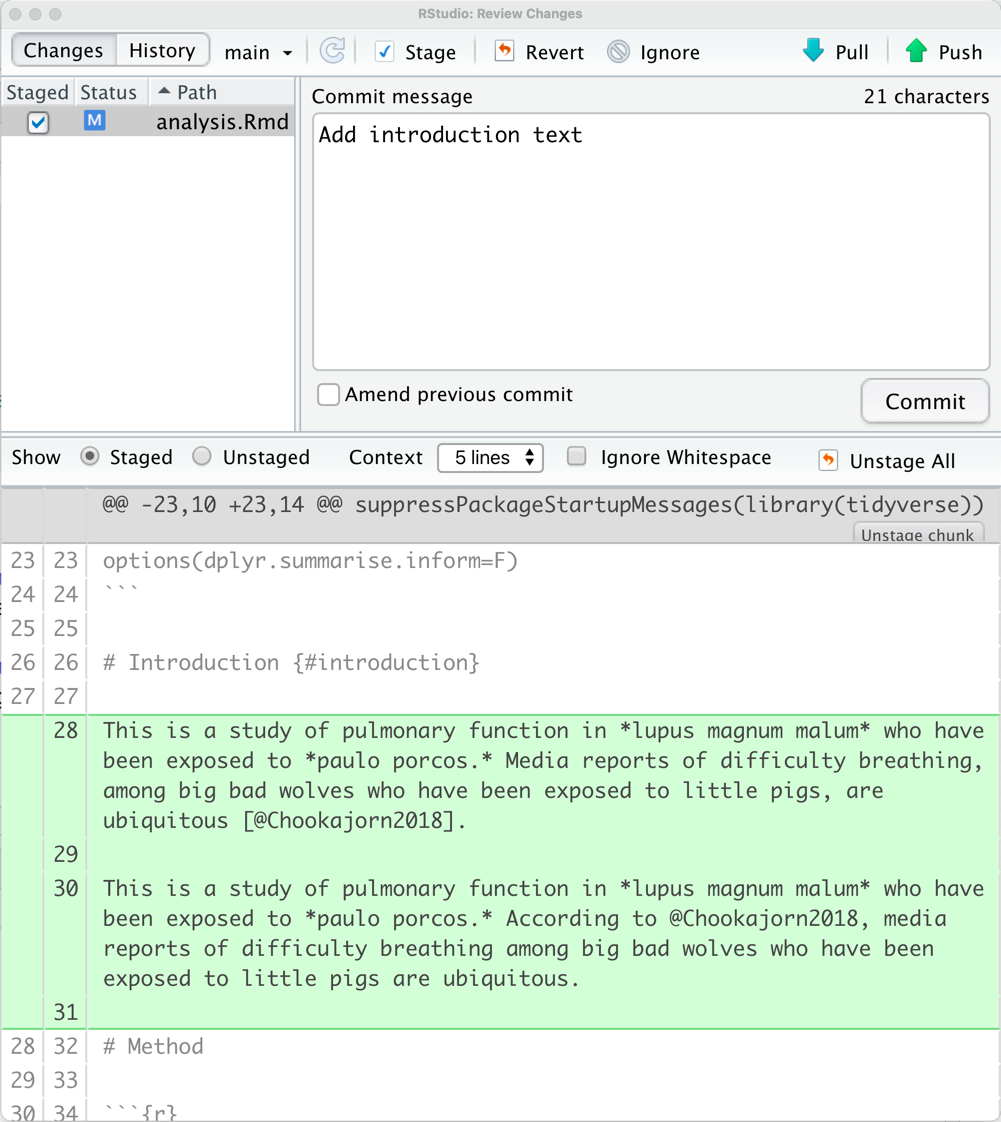
Dealing with Changes
To use the git terms:
- Save
- Stage (check off the box)
- Commit with a message (type a summary)
- Push (upload)
When you are on the commit screen you will see the sections that are changed. Deleted code will be in red. New code will be in green.
The GitHub Repo After Pushing Commits
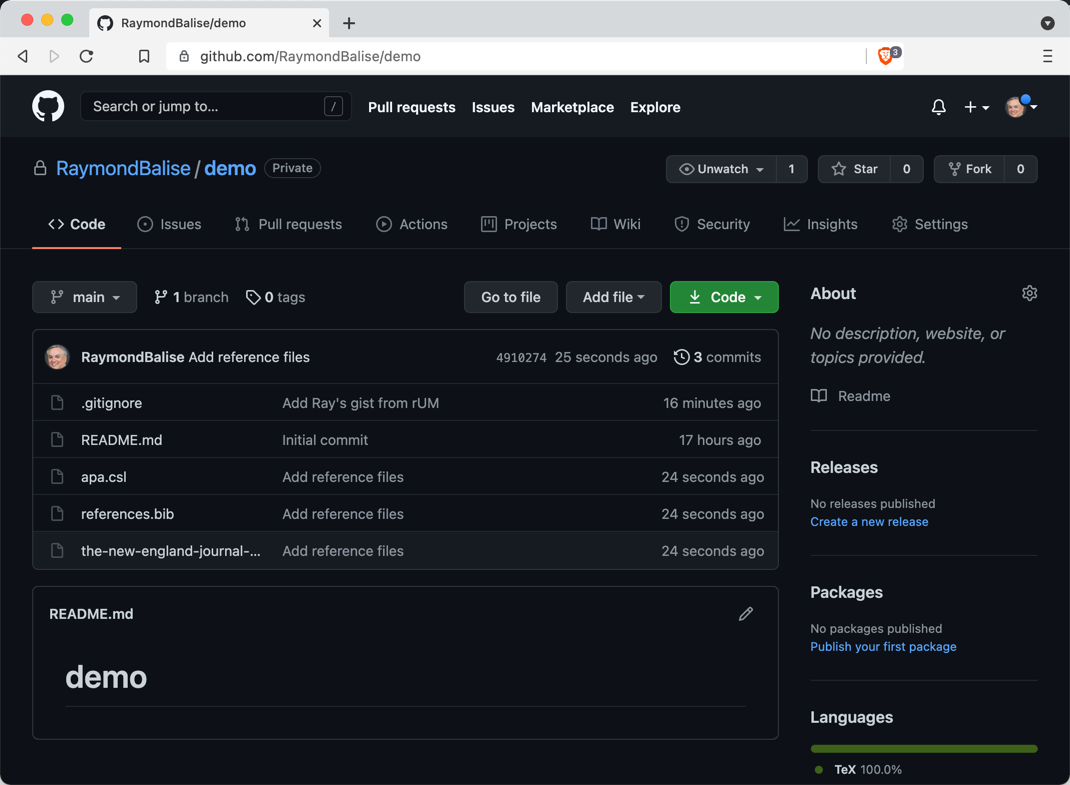
The GitHub Repo after Pushing Three Commits
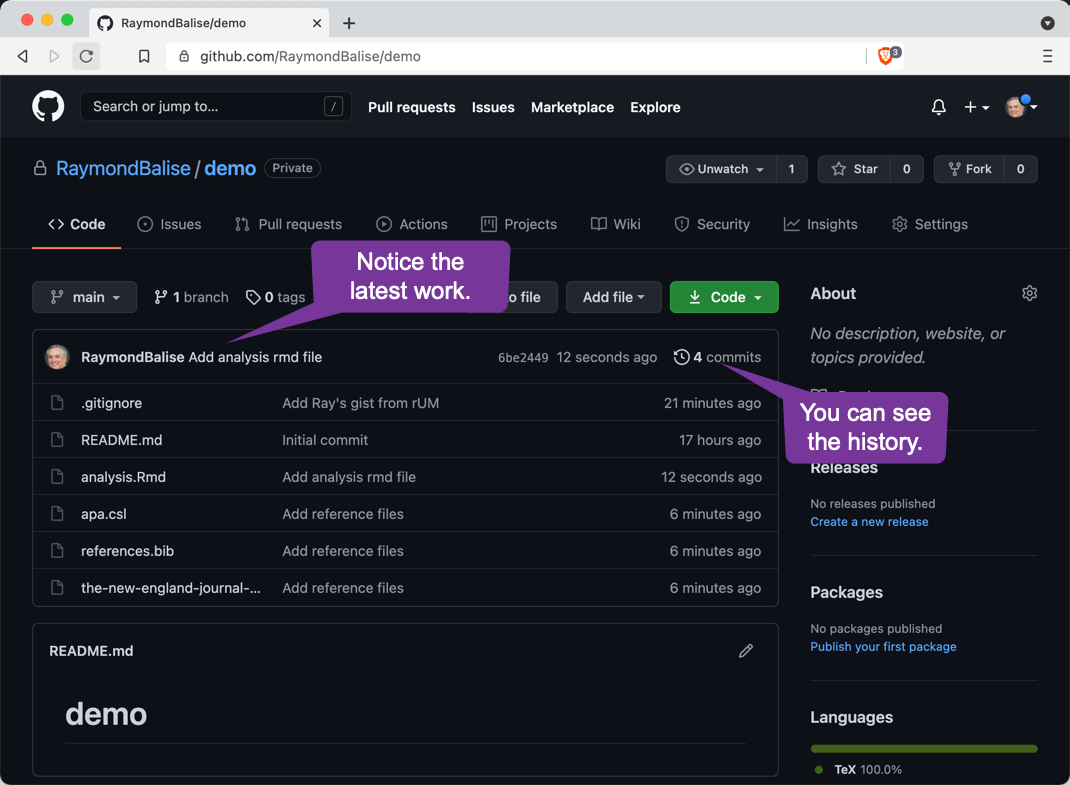
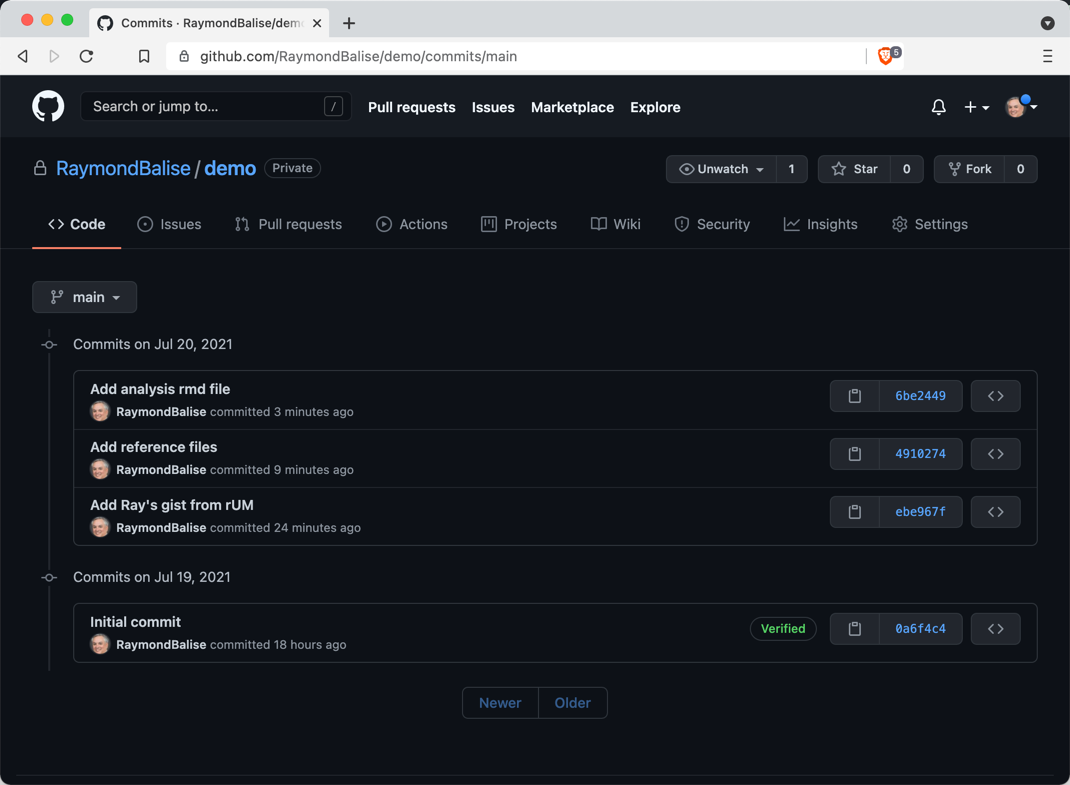
Commit History
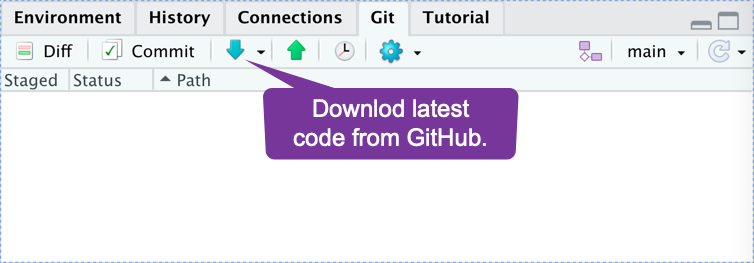
How do I get my project onto a new computer?
1) Follow the instructions to make a new GitHub project. That will give you the current state of the project.
2) Click the download button whenever you want to update the local information.
If You Want to Continue to Intimidate People
- Follow the instructions to make a new GitHub project. That will give you the current state of the project. Call this "cloning a project."
- Click the download button whenever you want to update the local information. Call this "pulling files."
Want to learn more about R Markdown?
- RStudio's website on R Markdown is excellent: https://rmarkdown.rstudio.com/
- Get the R Markdown cheat sheet: https://www.rstudio.com/wp-content/uploads/2015/02/rmarkdown-cheatsheet.pdf
- R Markdown: The Definitive Guide is a very readable manual and R Markdown Cookbook is superb for problem solving.


Want to learn more about git/GitHub?
The most popular reference for R git/GitHub integration is Happy Git and GitHub by Jenny Bryan and Jim Hester: https://happygitwithr.com/
Beyond the Basics
After you get comfortable with R Markdown, look at Emily Riederer's talk:
It is insightful and easy to follow.
Articles on Reproducible Research
Start here:
- Reproducible Research: A Retrospective by Roger Peng and Stephanie Hicks
Incredibly influential:
- 1,500 scientists lift the lid on reproducibility by Monya Baker
- What does research reproducibility mean? by Steven Goodman, Daniele Fanelli and John Ioannidis
Important:
- Statistical Analyses and Reproducible Research by Robert Gentleman and Duncan Temple Lang
- Repeatability of published microarray gene expression analyses by John Ioannidis et al.
- Reproducible research and Biostatistics by Roger Peng
- Reproducibility of computational workflows is automated using continuous analysis by Brett Beaulieu-Jones and Casey Greene
Coming in 2021
- I will "live code" a couple research projects using R and R Markdown and GitHub for CHARM in September 2021.